Good day everyone
I am trying to optimize the segments length for my lower extremity model (see pic below) using a static standing trial. However, after running the optimization, the foot and talus seem considerably small while the other segments' length seem reasonably optimized!
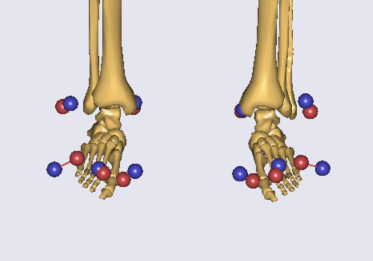
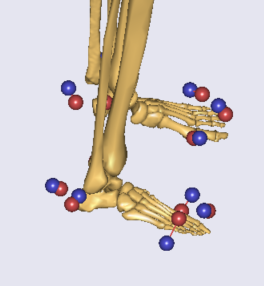
I do not know the cause of getting such small talus and foot. What could be the possible reasons?
-
As I understood, c3d markers are used to optimize the length of the segments. If that is true, how is this defined in AnyBody? Which markers does anybody identify to optimize the segment length?
1.1 Is using a static trial relevant to the issue? I found the following statement in Lesson 5 (Using Real motion data):
-
If the first question is true, Should I allow more of the foot markers to optimized? (which ones). would these markers, if they are optimized in position, represent a better foot dimension/ length. So, do I need to set Optx, Opty, Optz to On to optimize the length properly?
-
One last thing, in the model I also have done custom scaling/ morphing, and I excluded the talus and the foot? Is this relevant to the issue?
Regards,
Omar
Dear Omar,
-
It is correct that the markers are used to optimize the length of the segments in the parameter optimization and actually the static trial is a good one to determine the length of the segments. In the optimization, the model tries to minimize the objective function, so all the markers on the segment will be used if they are relevant to the parameter (the parameter to be optimized can be marker placement or length of the segments).
-
When optimizing the marker placement with Optx, Opty, Optz ON, you allow the marker (on the model not the data) to move and fit to the data marker. However, you should be careful if you make all of them ON, the segment will not catch a displacement or rotation, because the marker itself moves and the segment stays.
Small example: in the following picture with a segment and 1DOF rotation, if you allow the marker to move in all directions (Optx, Opty, Optz ON), the model marker will move exactly on top of the blue marker, and the segment does not rotate. But usually we want the segment to catch the movement and also optimize the marker placement (or segment length). So the best way is to have Optx, Opty, Optz OFF and Length_Opt ON or Optx=ON, Opty and Optz OFF.

And deciding which marker DOF you want to optimize, depends on the DOF you have in the model and DOF you want to optimize 
- About the third question, the answer is NO. Excluding them will not affect the optimization process

In the default model, in the LabSpecificData.any, you can see there is one DOF (foot length) to scale the whole foot (length, height, and width):
RIGHT_FOOT_LENGTH= ON,
LEFT_FOOT_LENGTH = ON,
So if we have Optx, Opty, Optz OFF for all the foot markers, we actually have one DOF to be optimized and it will use all the foot markers and constraints to do that.
I can see from the sagittal view that by optimizing the length of the foot, the overall 3 markers' locations are probably located in the optimal position and the model and data markers are close. So maybe you should take a look at the marker data you have. Maybe the markers in the model and in the data are placed a little bit differently. For example when they put the markers in the lab, they put it a bit posterior than the one defined in the model.
I hope this helps,
Hamed
Thank you Hamed for your response especially the example you provided  .
.
I still have some doubts and your explanation is appreciated.
-
You mentioned an objective function, what is it exactly? Is it the difference between model markers and the experimental markers? In this case, the model markers are created on the skeleton by sRelOpt. I can expect the accuracy of placing this marker on the skeleton to match the one from the c3d file may not always be a good placement  . Do you have a tip for this?
. Do you have a tip for this?
-
Does the optimization of the segment length (e.g., foot) try to bring the skeleton markers close to the experimental markers? thus, scaling/ optimizing the foot length?
-
To obtain the length of the foot, I can imagine that the heel marker and a marker on the tip of the 2nd toe will be necessary. In the figures above, I do have the heel marker but I do not have a marker on the tip of the 2nd toe. I rather have a marker at the proximal part of the 2nd toe proximal part (between mid and fore foot). Does this affect the optimization of the foot length?
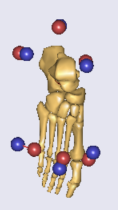
Because of the comma placed after "I do not think" I am a little bit confused haha. Can you clarify this please 
- As I mentioned before, I have a template model; I am not using the AMMR model; thus, LabSpecificData.any is not in my model. I checked the c3d file markers and I saw that the markers are placed properly at the same location where i defined the markers of the skeleton in the model. However, I would like to know if there is an extensive documentation for the parameter identification and how the segment length is optimized. I only found lesson 5 in the tutorials and it does not explain in depth the concept.
Best regards,
Omar
I am happy that the explanation was helpful 
-
Exactly as said. The optimization tries to minimize the difference between the model and experimental markers. And there are always inaccuracy between the marker placement. And that is one of the reasons that we can choose to optimize a marker placement according to experimental one (ny OptX, Y, and Z)  However, you should be careful. For example, if you choose to optimize all 3 markers on the anterior foot in X direction, then it cannot also optimize the length of the foot.
However, you should be careful. For example, if you choose to optimize all 3 markers on the anterior foot in X direction, then it cannot also optimize the length of the foot.
-
If the marker optimizations are OFF, then what you said is correct. It will try to minimize the distance of the markers by making the foot bigger and note that if you are using the default foot scaling, then the whole foot will scale by one factor and actually other markers on the foot will also affect the results. Because when the length of the foot is increased, the height and width of the foot will increase too and it may make a big gap between lateral markers even though the anterior markers look good  So the optimization will not find this solution because of the gap between lateral markers.
So the optimization will not find this solution because of the gap between lateral markers.
-
The best is that you have markers with the biggest distance to have less error. Here will be on the heel and the toe as you said. However, the optmizaiton will do exactly the same, and I think it is OK also with the markers you have.
-
Sorry it was a wrong comma. I meant, excluding them will not affect the optimization process.
-
Unfortunately not. But if you did not get the concept, you can continue asking here and other people with the same problem also will benefit that 
Best regards,
Hamed
Thank you Hamed for your response again.
I have another question:
If I can obtain the local coordinates of the markers with respect to the segment from the static trial and then create the marker on the skeleton based on those coordinates, is it better than manually creating it with sRelOpt?
I will give an example for illustration:
In the static trial I have Right Heel marker (RHeel), and I have already an algorithm that can calculate its local coordinates and save it in a file (Using AnyOutputFile). I then can use this file in the CreateMarkerDriver class to retrieve the marker's local coordinates and, of course, assign it to the segment (in this case the foot).
I now have some markers that are created with this method and the sRelOpt (manual) method. Is it better to have all the skeleton markers based on the new method?
I would like to hear your opinion on this cuz all I see in the tutorials and the webcasts is to use sRelOpt.
Best regards,
Omar
If you use the experimental data for marker placement on the model, then when you run it in the static trial, they will be at the same place. However, you cannot optimize the length of the foot because it will use the markers and because they are already at the minimum distance, then the scale factor will be 1. And you actually can do it by optimizing the markers in all directions. So they will end up at the same place as data. But this will not solve the foot length scaling.
Usually you define the marker placement with the anatomy. For example, when we have a maker in posterior calcaneus with the data, you should use the sRelOpt to move the marker on the model to the anatomical place of posterior calcaneus. And then the marker placement optimization should improve it.
Note that, the way you use the marker optimization is totally depends on how you trust your data. If you trust the data and know the exact anatomical place of it in the experimental data, you can use sRelOpt to move the marker to the anatomical place. Then you do not need to optmize the marker placement and you can achieve the foot length  You also can optimize two of the marker placements, and the other one will be used for the foot length.
You also can optimize two of the marker placements, and the other one will be used for the foot length.
BR,
Hamed