Hey, so I have R_FTC and L_FTC as a part of CAST markers in my c3d file for both Static and GIAT. Unfortunately, AnyBody does not have them in its inbuilt Marker Protocal file and I am trying to add them manually in the script with location as 'Most lateral prominence of the left/right greater trochanter' but I cannot find Trochanter, UpperFemure or GreaterFemoral etc in the 'Seg' file which I can use to put the markers on.
Please help.
Thank you.
Hi Romy,
Welcome to the AnyScript forum!
First of all, I would suggest you to go through this link to use your own data in the mocap model. It explains in general all the things that you should be aware of.
You can first of all explore the human leg. As you didn't find existing landmarks, you can create them yourself. Somewhere in your code, please write the following lines for example:
Main = {
Main.HumanModel.BodyModel.Right.Leg.Seg.Thigh = {
AnyRefNode Trochanter = {
sRel = {.....};
};
};
};
Alternatively, you could modify or create a copy of the existing markers in the marker protocol file to include your markers. In this way you can base the marker directly on the AnatomicalFrame by providing appropriate sRelOpt
// make sure to write #define _R_FTC in the beginning
#ifdef _R_FTC
// Marker on the Right Trochanter
CreateMarkerDriver R_FTC (
MarkerPlacement= Right.Leg.Seg.Thigh,
PlaceMarkerAt = AnatomicalFrame,
OptX=OFF,OptY=OFF,OptZ=OFF,
WeightX=1.0,WeightY=1.0,WeightZ=1.0,
UseC3DWeightResiduals=ON
) = {
sRelOpt = {0.0, -0.02, 0.05}; // approximately
};
#endif
Hope that helps.
Best regards,
Dave
Hey,
Thank you for the link. I will go through them! Your recommendations are well noted however, I have a small clarification: I tried to modify or create a copy of the existing markers in the marker protocal file to include the markers but I could not find the anatomical frame for the same and in order to introduce trochanter node in my Seg anyscript file I am confused with the sRel for the same!
I will go through the link now and get back if I find the solution.
Thank you.
Hi Romy,
Every segment in the human model has an anatomical frame. You should be able to find it if you explore to Main.HumanModel.BodyModel.Right.Leg.Seg.Thigh.AnatomicalFrame in the model tree (and likewise for other segments).
Regarding the sRel, this is just the relative location of the reference node to the parent reference frame, which can be the origin of a segment, another reference node, etc. You have two ways to create the marker at the right location: You either create the reference node on your human model and use this reference node in the marker protocol file, or you use another reference frame and provide the sRelOpt so that the marker is correctly located.
If you are confused about sRel, I would highly recommend that you go through the AnyBody tutorials. It is fundamental that you get an idea of the basics of AnyBody before you start working on detailed human model.
Also, to create a reference node on the leg, you don't necessarily need to make changes to the Seg file in the AMMR folder. I would advise against that as you will introduce the change in all the application examples using that copy of the AMMR. You can introduce changes to the body model through your specific model. The lines for making the reference node on thigh that I wrote in my previous post can be written in your model, for example in the LabSpecificData.any file. Moreover, I would suggest that you use the Scale function as is done for the rest of the reference nodes on the thigh segment (like you can see in the Seg file). The Scale function will make sure that the reference node is scaled according to the anthropometrics of the subject.
I hope that clarifies a bit more.
Best regards,
Dave
Hey,
I read through the suggested links and introduced the two markers in my 'MarkerProtocalStatic' file by using a existing anatomical mark in the thigh. However, I still have two clarifications.
- If I run the parameter identification with 1-2 Markers with Optimization=ON in just 1 direction to get a better experimental-body marker alignment. My body gets dismorphed. Is that common? Do we settle for the best we can achieve?
- I have the optimized parameters (Decent I hope). Since my experimental setup soley concentrates on Lower body I used Lowerextremity.main instead of MultiTrial StandingRef.main as opposed to the link you and AnyBody Tutorials says. Now I am getting errors while loading parameters from the above to MultiTrial FlySquat to run a dynamic simulation.
ERROR(SCR.EXP3) : ForcePlates.any(24) : Defined at : ForcePlateAutoDetection.any(112) : 'ForcePlateType' : Index [2] out of range for 'AnyInt[2]' I had no such errors with Forceplate when I ran in Lowerextremity.main. Can you help me understand why and how I can eradicate it?
Thank you.
Hi Romy,
- It really depends on the freedom in the optimization problem. Remember that also segment dimensions are getting optimized, so if you allow the vertical directions to be optimized as well as the segment dimension in case of the leg, then the solver can't find a good solution because all solutions are good.
- The LowerExtremity.main is from the Plug-in-gait_Simple model, if I understand correctly. These are two separate models with different settings for the lab. So, you will run into some errors like this becuase maybe the force plate settings are different, eg. number of force plates, forceplate type. You should stick to one model and work in that because then you will ensure you have consistent settings across your trials.
Best regards,
Dave
Hey again Dave,
I managed to get a decent parameter identification, thanks. I loaded the obtained parameters onto my GIAT and ran inverse dynamics which seems to be satisfactory! (By looking at the charts) These were to validate the c3d files and model.
I am now moving in to my next step which has my test subject laying in supine position on a bed placed on the force plates. Everything looked good until parameter identification. However, when I run the Inverse Dynamics (Using StandingRef) I get the following error message:
ERROR(SCR.EXP0) : JointsAndDriversOptimized.any(12) : UpdDriverPos : Error in evaluation. Please refer to the following error messages for details ...
ERROR(SCR.EXP.FUN2) : JointsAndDriversOptimized.any(12) : JntDriverTrunkFull : Error in function InterpolatedData : Called from object JntDriverTrunkFull : Parameter has an invalid value for this interpolation; extrapolations are not allowed.
Additional Information:
- I have added #define INCLUDE_UPPERBODY OFF in my TrialSpecifcData.any as I do not have any markers on my test subjects upper body as the experiment solely concentrates on lower body.
- As my Upperbody was not constrained I modified,
#define MOCAP_HUMAN_GROUND_RESIDUALS "PelvisTrunkAndContacts" //Changed.
in LabSpecificData.any by adding 'PelvisTrunkandContacts' in HumanGroundResiduals. In addition to Pelvis and Trunk, I have added 4 nodes (Thigh and Shank for both R&L) and 2 Heel to help the solver keep the body from falling.
When I skip the inverse dynamics and run 'FlywheelSquat' with parameters loaded from the StandingRef my Inverse dynamics run with the following message:
0.1.678) ...Inverse dynamic analysis completed
NOTICE(OBJ.OPR1) : AnyMocapModel.any(63) : InverseDynamicStudy.InverseDynamics : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
NOTICE(OBJ.OPR1) : RunApplication.any(52) : SubOperations : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
NOTICE(OBJ.OPR1) : RunApplication.any(49) : InverseDynamics : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
I would be happy to elaborate or give you additional details if necessary. Please help. Thanks in advance.
Hi Romy,
Regarding the error about interpolation... This happens because the joint angle output file, which has the results from marker tracking, contains data in a specific time period (start and end time of the trial), and when you tried to run inverse dynamics analysis, somehow, the inverse dynamics study is simulating another time period. For example, you may have changed the start time or end time of the trial and not run marker tracking again. The solution is to run marker tracking again.
I think, it's a little bit more confusing to understand what's happening in other trials. In one trial, you are simulating the subject lying on a bed, in another trial you are simulating the FlywheelSquat. These two trials clearly have different support conditions. The bed provides support all over the body, while in squat you are standing on your feet. So, you should be careful when you switch trials and ensure the right conditions are applied. The best way to do this is to make them in separate folders. I would also suggest you look at the seated human model to get an idea of how to simulate a bed. You could make the seat lie completely flat.
Best regards,
Dave
Greetings Dave,
Thank you, I figured that out and corrected them.
The points are noted but to shed more light on it: My test subject is laying flat (Supine position) with both ankles outside the end of bed frame.
A external load acts on the left ankle.
I am using StandingRef for parameter identification and use FlywheelSquat (with same C3D file as my test scenarios are static with just change in load acting on the ankle for different scenarios) so I have introduced 7 nodes in MyEnvironmentAdditions.any and used those nodes to modify my HumanGroundResiduals.any to have the following 6 constraints:
1 lin and 1 rot in trunk. (Exsisting trunk constraint)
1 lin in right shank node, left shank node, right thigh node, left thigh node.
I have not added heels as my ankle is placed outside the area of bed (hanging).
The JRF looks decent at the first glance but the HGR (for Lin LeftShank X and Y is a bit excessive) I assumed it must be the frictional force!
I am trying to introduce friction in it but reading your suggestion should I work on Seated human model or SpinePressureLyingBack so that the my model suits better?
Thank you.
Romy Jemals
Hi Romy,
Sorry for the delayed reply! It is a bit busy here in this period.
Yea, If you are working with a supine position, then the Seated Human or Spine Pressure Lying Back are great places to start with. In fact, I believe the Spine Pressure Lying Back model is basically the seated human model driven to the supine posture. This will make your work substantially easier. You would still need to modify some of the contact points as you mentioned the ankle are outside the bed frame.
Best regards,
Dave
Hey,
I understand, please don't be. ![]()
I took your advice and started working with Spine Pressure Lying Back but since it is a Validation model and not a MoCap working model.
I am curious if there is a possibility of me using the existing SPLB model with my c3d file and MoCap data?
I did try changing the body weight and height (curious to see results) but it leads to error as I presume a lot in the code is dependent on the above.
I am as of now working on StandingRef Mocap model but adapting it to resemble SPLB model by introducing Support.any, ConnectionsSegments.any, Environment.any, Initialpositions.any etc such as in the above.
I am not sure if it produces intended results but it is working. However, it's been a confusing ride so far as the coordinate systems between my model and SPLB model are different and certain code sections like sRel,ARel (for SeatPelvisJntNode, PelvisSeatJntNode,SeatAdjNode, etc) are not explained in the SPLB model: As in why certain nodes need to be in same coordinate axis and why a few are not. It would be beneficial if I could understand the underlying concept.
Thank you.
With Regards,
Romy Jemals
Hi Romy,
You will need to port the seat/bed model to your own mocap model. This means you will have to bring in the code for segments, support, joints and drivers for the seat. I think that's exactly what you have described, so it should be alright. It will be a bit of work, but since you are working with marker based data, you will only have to do this one time. You basically need to position the bed in a sensible way according to your mocap data. This can of course be complicated for a whole bunch of reasons. Most critical, as you say, would be if your lab coordinate system is not consistent with the global ref in the SPLB validation model. I think, you should be able to adjust some of the input parameters or drivers between the seat and the global ref to account for that. The rest of the segments (backrest, footrest, etc) should simply follow the corresponding drivers between the seat and the segment.
Best regards,
Dave
Greetings Dave,
Thank you for your timely response. All the said points are noted. However, I have a remark and a question to follow.
Remark being: Since my test scenario is static case, I have skipped drivers. I have instead used AnyKinEq for BedRest, BackRest, HeadRest, LegRest with the following:
MeasureOrganizer = {0, 1, 2, 3, 4, 5}; and
Type = RotVector; for AnyKinRotational.
Is that okay?
- I am the verge of completing my model to resemble SPLB Model but I have not modified my ARel for any node so far and let it in its default position. Do you reckon I fix it like the SPLB model for complete validation/success or I leave it in their own default coordinate axes. Incase yes, Can I know why certain nodes need to be in same coordinate axis and why a few should not. It would be beneficial if I could understand the underlying concept to replicate.
Thank you.
With Regards,
Romy Jemals
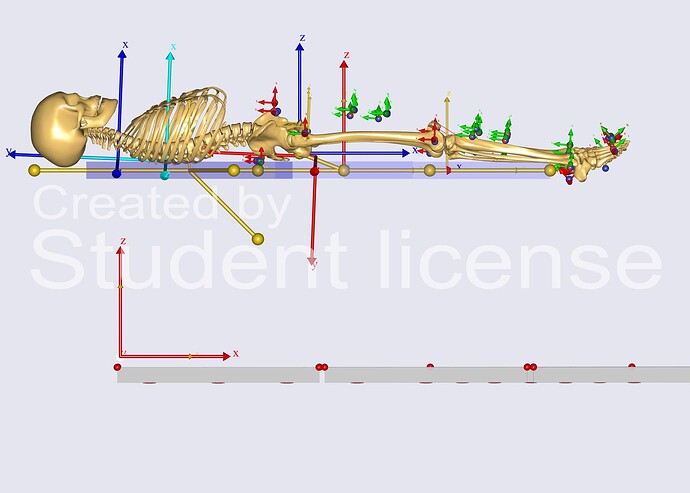
In addition:
I just ran the Inverse Dynamics study. ParameterIdentification and Marker Tracking work fine. The body is placed on the bed segment (as close as possible to SPLB model except the coordinate axes alignment).
This is how it looks after Marker Tracking.
However, when I run InverseDynamics I get the following error:
- Operation Sequence: (Operation: Main.RunAnalysis.InverseDynamics):
0.0) Operation Sequence: (Operation: Main.RunAnalysis.InverseDynamics.SubOperations):
0.0.0) Dummy operation: (Operation: Main.RunAnalysis.InverseDynamics.SubOperations.dummy):
0.1.0) InverseDynamics (Operation: Main.Studies.InverseDynamicStudy.InverseDynamics):
0.1.0.0) PreOperation (Operation: Main.Studies.InverseDynamicStudy.InverseDynamics.PreOperation):
0.1.0.0.0) InitialConditions (Operation: Main.Studies.InverseDynamicStudy.InitialConditions):
0.1.0.0.0) ...Design variables have been updated.
0.1.0.0.1) ...Load-time positions have been re-established.
0.1.0.0.1.0) PreOperation (Operation: Main.Studies.InverseDynamicStudy.InitialConditions.PreOperation):
0.1.0.0.1.0.0) Operation Sequence: (Operation: Main.ModelSetup.Views.SetViewMacros.InverseDynamicView):
0.1.0.0.1.0.0.0) mcr (Operation: Main.ModelSetup.Views.SetViewMacros.InverseDynamicView.mcr):
Macro command : mcr(1:1)> classoperation Main.ModelSetup.Views.InverseDynamicView"Set View"
0.1.0.0.2) ...Kinematic analysis completed.
0.1.0.0.3) ...Dependent variables are fully updated.
0.1.0) Inverse dynamic analysis...
ERROR(OBJ.MCH.MUS4) : AnyMocapModel.any(63) : InverseDynamicStudy.InverseDynamics : Muscle recruitment solver : solver aborted due to singular KKT matrix
Can you help me understand why I get this and how I should proceed to eradicate the said error?
Thank you.
With Regards,
Romy Jemals
Hi Romy,
It's great to see you are making progress. It seems you have the bed correctly located with respect to the subject ![]()
Regarding your previous post, it doesn't matter whether you use AnyKinEq, or drivers. They are just different ways in which you can constrain the kinematics of the bed, and fundamentally they are the same. You can think of AnyKinEq as a driver that is driven to zero. The important thing is that everything is in the right place, and it doesn't matter so much how you drive it to the right place.
Likewise, for ARel, it doesn't matter so much. The only underlying concept is that you want to have a scheme that you follow when you make a model and then make the joints accordingly. When you have to define joints or drivers, you have to be aware of the axis, so it's all interrelated. It's simply a choice that the user makes while modelling and it shouldn't matter so much. When you try to make a complex model, linking multiple segments, normally you would have a "root" segment (like the pelvis for the human model) that is driven with respect to the global ref, and everything else will follow.
Coming to the error in inverse dynamics, this is normally the case when your model doesn't have enough support to balance all the degrees of freedom. In other words, you are missing some reaction force, or muscles, to balance the model. I have a couple of questions for you:
- Have you brought the code defining the support between the human model and the bed to your model? It will create some reference nodes on the human model and define conditional contact using recruited actuators.
- When you made the kinematic constraints to fix the bed, are these kinematic constraints to the global ref? Do they have their Reaction.Type set to On? This will allow reaction forces between the bed and the global ref.
Best regards,
Dave
Greetings Dave,
I apologize for getting back late. I was trying to implement your suggestions to eradicate the error. To sum up my timeline thus far:
- I could not eradicate the singular KKT matrix error but I ran it in different computer (out of curiosity) and noticed the ID study runs without any errors! Unfortunately, The model-bed setup did not yield acceptable results. Hip, Knee forces were in magnitude > 500 N. (Not all but a few & Hip PD for both legs > 1000 N)
To answer your questions: - Yes, I did bring the code defining the support between the human model and bed to my model.
- The Kinematic constraints are to global ref, as far I know. The reaction type is set to ON.
Can you explain why it throws error in one and runs in another and how I can fix the unrealistic forces?
Please let me know if you need additional information from my end to explain the question.
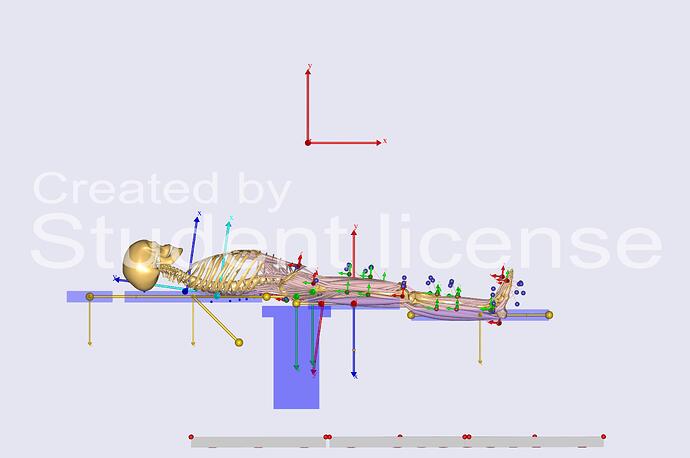
- Since I could not find a way around the above issue and as I noticed the slight pelvis tilt through the bed during the Inverse dynamics study and realized the model by default was not similar to the validation model (in terms of model setup and position) so I set up the bed inclined to support my model's natural supine position and also set up the coordinate axis to resemble the SPLB model and this is how it looked,
However, when I run it now I get the following:
- Operation Sequence: (Operation: Main.RunAnalysis):
0.0) Operation Sequence: (Operation: Main.RunAnalysis.LoadParameters):
0.0.0) Operation Sequence: (Operation: Main.RunAnalysis.LoadParameters.SubOperations):
0.0.0.0) Dummy operation: (Operation: Main.RunAnalysis.LoadParameters.SubOperations.dummy):
0.1.0) Load_parameters (Operation: Main.ModelSetup.Macros.Load_parameters):
Macro command : Load_parameters(1:10)> classoperation Main "Load Values" --file="\filur02.it.liu.se\students\romdo288\Documents\AnyBody.8.0.x\AMMR.v3.0.4-Demo\Application\MocapExamples\Plug-in-gait_MultiTrial_StandingRef\Output/S4_StandingRef.anyset"
Loading and overriding values from : '\filur02.it.liu.se\students\romdo288\Documents\AnyBody.8.0.x\AMMR.v3.0.4-Demo\Application\MocapExamples\Plug-in-gait_MultiTrial_StandingRef\Output\S4_StandingRef.anyset' ...
...Values updated
Macro command : Load_parameters(10:10)> classoperation Main "Update Values"
Updating expressions...
WARNING for Main.EnvironmentModel.EnvironmentModel.GlobalRef: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.BackRest: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.Seat: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.HeadRest: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.LegRest: Non-orthogonal axes were adjusted.
...Finished updating expressions
1.0) Operation Sequence: (Operation: Main.RunAnalysis.MarkerTracking):
1.0.0) Operation Sequence: (Operation: Main.RunAnalysis.MarkerTracking.SubOperations):
1.0.0.0) Dummy operation: (Operation: Main.RunAnalysis.MarkerTracking.SubOperations.dummy):
1.0.1.0) Kinematics (Operation: Main.Studies.MarkerTracking.Kinematics):
1.0.1.0.0) PreOperation (Operation: Main.Studies.MarkerTracking.Kinematics.PreOperation):
1.0.1.0.0.0) InitialConditions (Operation: Main.Studies.MarkerTracking.InitialConditions):
WARNING for Main.EnvironmentModel.EnvironmentModel.GlobalRef: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.BackRest: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.Seat: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.HeadRest: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.LegRest: Non-orthogonal axes were adjusted.
1.0.1.0.0.0) ...Design variables have been updated.
1.0.1.0.0.1) ...Load-time positions have been re-established.
1.0.1.0.0.1.0) PreOperation (Operation: Main.Studies.MarkerTracking.InitialConditions.PreOperation):
1.0.1.0.0.1.0.0) Operation Sequence: (Operation: Main.ModelSetup.Views.SetViewMacros.KinematicView):
1.0.1.0.0.1.0.0.0) mcr (Operation: Main.ModelSetup.Views.SetViewMacros.KinematicView.mcr):
Macro command : mcr(1:1)> classoperation Main.ModelSetup.Views.KinematicView"Set View"
1.0.1.0.0.2) ...Kinematic analysis completed.
1.0.1.0.0.3) ...Dependent variables are fully updated.
1.0.1.0) Kinematic analysis...
1.0.1.641) ...Kinematic analysis completed
2.0) Operation Sequence: (Operation: Main.RunAnalysis.InverseDynamics):
2.0.0) Operation Sequence: (Operation: Main.RunAnalysis.InverseDynamics.SubOperations):
2.0.0.0) Dummy operation: (Operation: Main.RunAnalysis.InverseDynamics.SubOperations.dummy):
2.0.1.0) InverseDynamics (Operation: Main.Studies.InverseDynamicStudy.InverseDynamics):
2.0.1.0.0) PreOperation (Operation: Main.Studies.InverseDynamicStudy.InverseDynamics.PreOperation):
2.0.1.0.0.0) InitialConditions (Operation: Main.Studies.InverseDynamicStudy.InitialConditions):
WARNING for Main.EnvironmentModel.EnvironmentModel.GlobalRef: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.BackRest: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.Seat: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.HeadRest: Non-orthogonal axes were adjusted.
WARNING for Main.EnvironmentModel.EnvironmentModel.Seg.LegRest: Non-orthogonal axes were adjusted.
2.0.1.0.0.0) ...Design variables have been updated.
2.0.1.0.0.1) ...Load-time positions have been re-established.
2.0.1.0.0.1.0) PreOperation (Operation: Main.Studies.InverseDynamicStudy.InitialConditions.PreOperation):
2.0.1.0.0.1.0.0) Operation Sequence: (Operation: Main.ModelSetup.Views.SetViewMacros.InverseDynamicView):
2.0.1.0.0.1.0.0.0) mcr (Operation: Main.ModelSetup.Views.SetViewMacros.InverseDynamicView.mcr):
Macro command : mcr(1:1)> classoperation Main.ModelSetup.Views.InverseDynamicView"Set View"
Constraint violations for study 'Main.Studies.InverseDynamicStudy' :
ERROR(OBJ.MCH.KIN3) : AnyMocapModel.any(63) : InverseDynamicStudy.InitialConditions : Kinematic analysis failed in time step 0 : Velocity analysis is not completed
NOTICE(OBJ.OPR1) : AnyMocapModel.any(63) : InverseDynamicStudy.InitialConditions : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
NOTICE(OBJ.OPR1) : AnyMocapModel.any(63) : InverseDynamicStudy.InverseDynamics.PreOperation : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
2.0.1.0) Inverse dynamic analysis...
Constraint violations for study 'Main.Studies.InverseDynamicStudy' :
Constraint #246 is above tolerance 1e-05, error = 0.007743, constr. #0 in 'Main.EnvironmentModel.EnvironmentModel.Jnt.BackRestHeadRestJnt.Constraints'.
Constraint #247 is above tolerance 1e-05, error = 0.001029, constr. #1 in 'Main.EnvironmentModel.EnvironmentModel.Jnt.BackRestHeadRestJnt.Constraints'.
Constraint #251 is above tolerance 1e-05, error = 0.000011, constr. #0 in 'Main.EnvironmentModel.EnvironmentModel.Jnt.SeatLegRestJnt.Constraints'.
Constraint #252 is above tolerance 1e-05, error = 0.000108, constr. #1 in 'Main.EnvironmentModel.EnvironmentModel.Jnt.SeatLegRestJnt.Constraints'.
Constraint #253 is above tolerance 1e-05, error = 0.000039, constr. #2 in 'Main.EnvironmentModel.EnvironmentModel.Jnt.SeatLegRestJnt.Constraints'.
Constraint #255 is above tolerance 1e-05, error = 0.000020, constr. #4 in 'Main.EnvironmentModel.EnvironmentModel.Jnt.SeatLegRestJnt.Constraints'.
Constraint #379 is above tolerance 1e-05, error = 0.000148, segment constr. 'Main.EnvironmentModel.EnvironmentModel.Seg.LegRest'.
ERROR(OBJ.MCH.KIN3) : AnyMocapModel.any(63) : InverseDynamicStudy.Kinematics : Kinematic analysis failed in time step 0 : Failed to solve position constraints
2.0.1.0) ...Inverse dynamic analysis terminated
NOTICE(OBJ.OPR1) : AnyMocapModel.any(63) : InverseDynamicStudy.InverseDynamics : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
NOTICE(OBJ.OPR1) : RunApplication.any(52) : SubOperations : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
NOTICE(OBJ.OPR1) : RunApplication.any(49) : InverseDynamics : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
3.0) Operation Sequence: (Operation: Main.RunAnalysis.SaveOutput):
3.0.0) SaveOutputToHDF (Operation: Main.ModelSetup.Macros.SaveOutputToHDF):
Macro command : SaveOutputToHDF(1:2)> classoperation Main.Studies.MarkerTracking.Output"Save data"--type= "Deep"--file= "\filur02.it.liu.se\students\romdo288\Documents\AnyBody.8.0.x\AMMR.v3.0.4-Demo\Application\MocapExamples\Plug-in-gait_MultiTrial_StandingRef\Output/S4_FlywheelSquat_MarkerTracking.anydata.h5"
Main.Studies.MarkerTracking.Output : Saving output data file...
...saving succeed.
Elapsed Time :61.406
Macro command : SaveOutputToHDF(2:2)> classoperation Main.Studies.InverseDynamicStudy.Output"Save data"--type= "Deep"--file= "\filur02.it.liu.se\students\romdo288\Documents\AnyBody.8.0.x\AMMR.v3.0.4-Demo\Application\MocapExamples\Plug-in-gait_MultiTrial_StandingRef\Output/S4_FlywheelSquat_InverseDynamicStudy.anydata.h5"
Main.Studies.InverseDynamicStudy.Output : Saving output data file...
...saving succeed.
Elapsed Time :76.266
NOTICE(OBJ.OPR1) : RunApplication.any(33) : RunAnalysis : Operation finished with non-fatal errors. Please verify whether these errors are acceptable.
Please let me know how I can work around the issue.
Thank you.
Kind Regards,
Romy Jemals
In addition to that, I have been thinking about alternative to this (As it is consuming time) and I would like to know how I can adapt the existing SPLB model to my test subject (Body height and mass). I already figured to induce the load on the left ankle and the mass but if I change the height, the ID study collapses as the bed segments are dependent on the model height I presume. How do I come around this issue?
Thank you.
With regards,
Romy Jemals.
Hi Romy,
It has been a long time indeed and I may end up saying things that contradict something I said earlier, because I am not so fresh with the topic.
It is indeed very strange why you are able to run the model on one computer and not another. The only explanation I can think of is a difference in AMS or AMMR versions. If it is all the same, then it is definitely not something I can explain. You should get the same results, irrespective of the machine.
Your model looked like it was ok in your previous posts, so I am also a bit confused what exactly is not working. At this point, I must mention some other basic checks:
is the gravity acting in the correct direction? Since you are seeing such large forces, I am starting to wonder about the gravity direction.
Either that, or the contact conditions between the human and bed.
Regarding the contact between the human and the bed, can you just run the kinematics to check that the contact points are within the contact threshold? Also, have you set them up correctly for the direction? Is the normal direction correctly specified for the contact?
Are the bed model and all the constraints/reactions correctly included in the inverse dynamic study?
One last thing I would think about is the force plates. Are they used somewhere in the model? If yes, how? If no, maybe it's best to remove them.
When I look at the image where you have tried to adjust the bed incline, I am not sure what to conclude, because the legs are way off from the markers. So it seems, you have some kinematic constraints that are keeping the bed and the human stuck together. This is likely going to cause an error at some point. You should think about this a little bit. What is driving the posture of the human, is it the markers or the bed? My answer would be the markers, and in this case, the bed should be constrained in a way so that it follows where the human is. Alternatively, the bed could simply be a static object that is fixed in space and does not need to be adjusted.
I am also concerned about the warnings regarding Non-orthogonal axes were adjusted. It looks like you have specified the axes of the bed segments incorrectly. In AnyBody, we use the right-hand cartesian coordinate system. Possibly, some of the axes you have defined are not following this system.
Lastly, you can surely adjust the existing SPLB model in the AMMR. You can update the anthropometry of the subject through the AnyManUniform file, but at the same time, you will need to adjust the drivers of the seat.
Best regards,
Dave
Greetings,
Thanks for your detailed feedback! I’ve gone through all the points you raised, and here’s my response:
- Yes, I have included the necessary support definitions in my model by using: #include "ConnectionSegments.any" #include "Support.any". Within
Support.any, I have included:
BackrestSupport.any
SeatSupport.any
LegrestSupport.any
I skipped Ulna, ArmRest, and FootRest because: My model does not have arms, so Ulna and ArmRest are not relevant.The **heel needs to hang outside the bed, so I did not include FootRest support.
-
The kinematic constraints to fix the bed are applied to the global reference frame.
The Reaction.Type is set to {On, On, On, On, On, On}, which should allow reaction forces between the bed and the global reference. -
I also find this strange. The AMS and AMMR versions should be the same on both machines, but I will double-check to confirm.
-
Previous Model:
i) Gravity is currently set as Gravity = {0.0, 0.0 , -9.81}; , as the Z-axis is pointing upward in my coordinate system and this aligns with my C3D file coordinate systems as well.
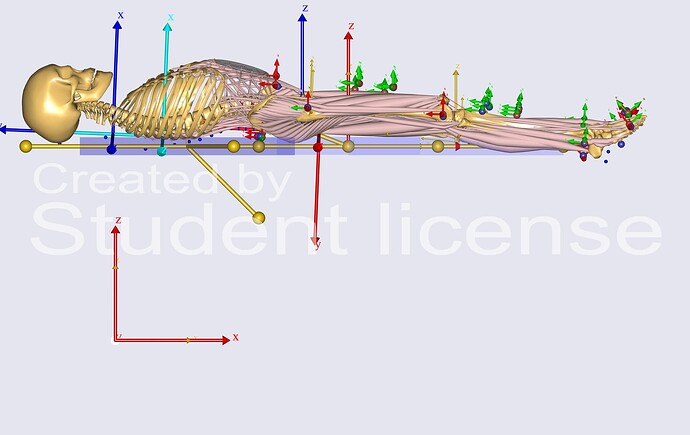
ii) Regarding the contact between the human and the bed, I ran load parameters, marker tracking, initial conditions, kinematics and they all worked fine. However, I am unsure if the contact points are within the contact threshold. The contact setup follows default directions, but I did modify Axes0 for the segment to suit my model. ARel was mostly unchanged.
I am attaching the image below for your reference.
iii) The bed model and all constraints/reactions are included in the inverse dynamics study to my knowledge. I have not made any changes to the existing ID study code which is,
/// Study for running the inverse dynamic simulation
AnyBodyStudy InverseDynamicStudy =
{
AnyComponentDefinition obj = {};
Gravity = Main.ModelSetup.LabSpecificData.Gravity;
AnyFolder& C3DData= Main.ModelSetup.C3DFileData ;
AnyFolder& BodyModel= Main.HumanModel.BodyModel;
AnyFolder& EnvironmentModel = Main.EnvironmentModel;
AnyFolder ModelEnvironmentConnection =
{
#include "HumanGroundResiduals.any"
#include "JointsAndDriversOptimized.any"
};
// Include markers for visualization, but exclude the driver part
AnyFolder &Markers= Main.ModelSetup.MocapDrivers;
// Exclude marker drivers and soft rhythms
#include "ExcludeSoftDrivers.any"
#include "InverseDynamicSettings.any"
};
Should I include the following as in SPLB model?
//Size of total force in the L5 Sacrum joint
AnyVar L5SacrumReac = (Main.Model.BodyModel.Trunk.JointsLumbar.L5SacrumJnt.Constraints.Reaction.Fout[0]^2+
Main.Model.BodyModel.Trunk.JointsLumbar.L5SacrumJnt.Constraints.Reaction.Fout[1]^2+
Main.Model.BodyModel.Trunk.JointsLumbar.L5SacrumJnt.Constraints.Reaction.Fout[2]^2)^0.5;
Or Do you mean anything else?
iv) Yes, the force plates are present in model and I assume they are active for force calculations. I ran the ID study by removing them but it still does not work.
Object description:
Total number of rigid-body d.o.f.: 288
Total number of joint coordinates: 80
Total number of constraints:
Joints: 160
Drivers: 98
Other: 30
Total: 288
Total number of active reaction and driver forces: 221
I still get the following error message in my system:
- InverseDynamics (Operation: Main.Studies.InverseDynamicStudy.InverseDynamics):
0.0) PreOperation (Operation: Main.Studies.InverseDynamicStudy.InverseDynamics.PreOperation):
0.0.0) InitialConditions (Operation: Main.Studies.InverseDynamicStudy.InitialConditions):
0.0.0) ...Design variables have been updated.
0.0.1) ...Load-time positions have been re-established.
0.0.1.0) PreOperation (Operation: Main.Studies.InverseDynamicStudy.InitialConditions.PreOperation):
0.0.1.0.0) Operation Sequence: (Operation: Main.ModelSetup.Views.SetViewMacros.InverseDynamicView):
0.0.1.0.0.0) mcr (Operation: Main.ModelSetup.Views.SetViewMacros.InverseDynamicView.mcr):
Macro command : mcr(1:1)> classoperation Main.ModelSetup.Views.InverseDynamicView"Set View"
0.0.2) ...Kinematic analysis completed.
0.0.3) ...Dependent variables are fully updated.
0) Inverse dynamic analysis...
ERROR(OBJ.MCH.MUS4) : AnyMocapModel.any(63) : InverseDynamicStudy.InverseDynamics : Muscle recruitment solver : solver aborted due to singular KKT matrix
And in other system, it runs fine with unlikely large forces.
New model:
Regarding the inclined bed model, It was me acting out of desperation to figure out a solution by thinking it would solve the problem and the warnings regarding Non-orthogonal axes were adjusted are because I tried to duplicate my new model to SPLB model by changing the node coordinates etc. However, We do not have to dwell more into this.
I'd be content if we could solve the problem with the previous model.
Lastly, to adjust the existing SPLB model in the AMMR. I did update the anthropometry of the subject through the AnyManUniform file, but at the same time, I could not figure out which how to adjust the drivers of the seat but I will take a look at them now.
Thank you.
With Regards,
Romy Jemals
Hi Romy,
I think we should focus on the human-bed contact setup. From the image, it looks like the leg might be out of the contact threshold.
Can you increase the threshold distance for contact?
Also, the normal direction is critical since this code tries to simulate contact force.
You can find the settings in files for support (say, SeatSupport.any). Please check and increase UserDefinedLimitHigh. And also check the variable Direction. It is the base object that determines the normal direction.
Are all your bed segments (backrest, seat, legrest) individually fixed to the global ref? With reactions set to On?
Best regards
Dave