Hello AnyBody Community,
- AnyBody.8.1.4 (only recently updated the software from 8.0.0)
- AMMR4-Beta
- Plug-in-gait_MultiTrial_StandingRef with flexible thoracic model, incorporating simple muscles+ligaments+non linear disc+soft rhythm for all 3 spine region
- Driven by mocap data of several dozens of subjects, reflecting ADL movements
- Batch process with Python/Jupiter Notebook
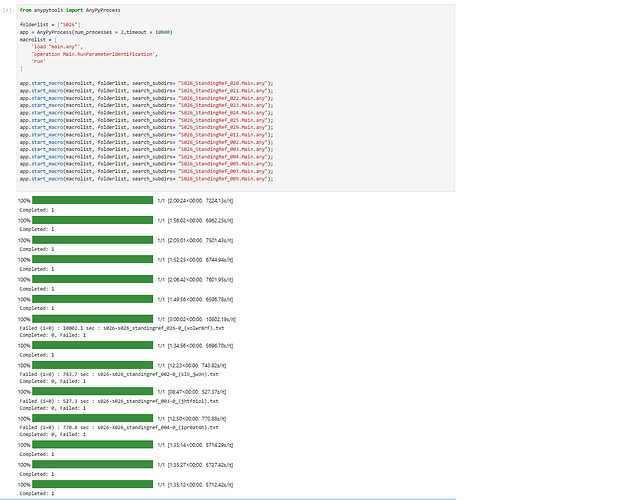
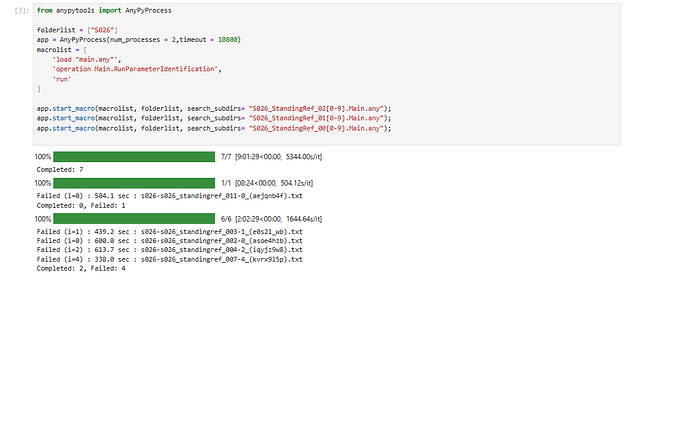
As usual, I would run Parameter Identification for every subject that I have collected mocap data from. As there is no absolute minimum requirement of movement needed for C3D file to be used to run the ParamIdent, I randomly choose one file from a walking trial, as the example shown in the tutorial. However, I frequently encountered that the ParamIdent has failed and I had to choose a different C3D file. Sometimes, I had to repeat this more than twice for the same subject as the ParamIdent had failed in both times. So I decided just run all of the C3D trial files (for all the ADL involving standing position) I have (for that particular subject) with batch processing to prevent anymore frustration and wasting more time. Then I would choose one of the successful ParamIdent outputs (ANYSET file) as the scaling and marker parameter reference file for actual Kin/InvDyn simulations.
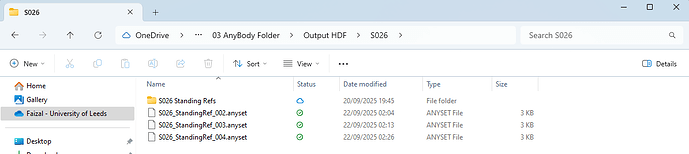
Nonetheless, my strategy to use batch processing the ParamIdent is suddenly became flawed when output from successful simulation is not being saved (at all) but any failed one is 'saved'. I am not sure if the issue is because I ran ParamIdent with batch processing or Python/Jupiter code itself or AnyBody con/version.I tried to code all the ParamIdent simulation as individual files as well as a group but both still not saving the Anyset file for any successful simulation. See photos below.
Let me know what you all think...
Thank you.
Kind regards,
Faizal